Genomic data management research at politecnico di milano




NGS
In the coming decade, Next Generation Sequencing (NGS) will offer a fast and inexpensive technology (few hours and few hundreds of dollars) to read the whole human genome.
The data management community is not ready for NGS: data are managed by a variety of tools focused on specific data extractions and transformations, with specific physical formats and no focus on interoperability.
![]()
The human genome is a sequence of 3 billions of dna nucleotides.
The potential for their data querying, analysis and sharing may be considered as the biggest and most important "big data problem" of mankind.
GMQL
A new holistic and abstract approach that uses cloud computing technologies and an interoperable data format to store and query tens of datasets, thousands of samples and several millions of DNA regions in order to discover interesting dna regions and their relationships. Thanks to algebraic operations on both DNA regions and metadata, GMQL genome-wide queries are able to find interesting regions by combining mutations, expression or regulation experiments
GMQL
![]()
![]()
![]()
GMQL supports efficient high-level query processing of thousands of experimental data samples, produced with a variety of experimental methods and encoded in a variety of data formats, together with their biological and clinical metadata descriptions as well as multiple annotation data. It supports big data analysis combining hundreds of samples with millions of regions.
Current tools for “big data” (e.g. BioPig, GQL) operate on reads and typically operate “sample by sample”. Other tools which are used to combine samples and working on their regions (e.g. BEDTools, BEDOPS) operate only on bed files - with a “scripting style” of programming.
GMQL instead, focused on assisting knowledge extraction, is meant to operate on higher level data obtained after raw data preprocessing and feature calling, rather than on raw data directly. This offers the advantage of not interfering with the variety of data preprocessing tools and pipelines that are already in place in the different research centers, as well as of directly benefiting of their output, thanks to the interoperability and data integration support.
Example: Identification of distal bindings in transcription regulatory regions
Public data can be used not only by themselves, but also together with in house produced experimental data, for integrated evaluations and comparisons with increased support. GMQL allows a powerful genome-wide processing and leverages the high-level data, including variant calling, gene expression and region enrichment (i.e. peak) calling data, which are increasingly available within public large data collections (e.g. 1000 Genomes Project, TCGA, and ENCODE).
Genomic data model (GDM)
Our Genomic Data model (GDM) provides abstractions for DNA regions of the sample and for metadata describing the sample's properties for a simplified structured outcome and ideal format for data analysis. MAP operations, through reference region R, extract and standardize genomic features expressed in distinct dataset.
![]()
Genometric query system architecture
Architecture operating upon cloud computing systems based on Hadoop and operating on four different layers: services, orchestration & translation, engines , management of files and data sources and including several integrated components for data management.
for queries submitting, their execution monitoring and results retrieving.
submitting PIG instructions with appropriate settings so as to optimize performances.
providing metadata indexing using Lucene.
translating GMQL code to Pig Apache scripts.
supporting the ad-hoc transformation of data from native formats to GDM after sample selection through indexing.
![]()
Genome space analysis
We are studying user-friendly interfaces to further facilitate the user interaction with our system to express GMQL queries and to search genomic data patterns of interest by visually drawing them in a genome browser. We developed an input tool for browsing metadata, which anticipates the effect of sample selections, so as to visualize the number and metadata of selected samples.
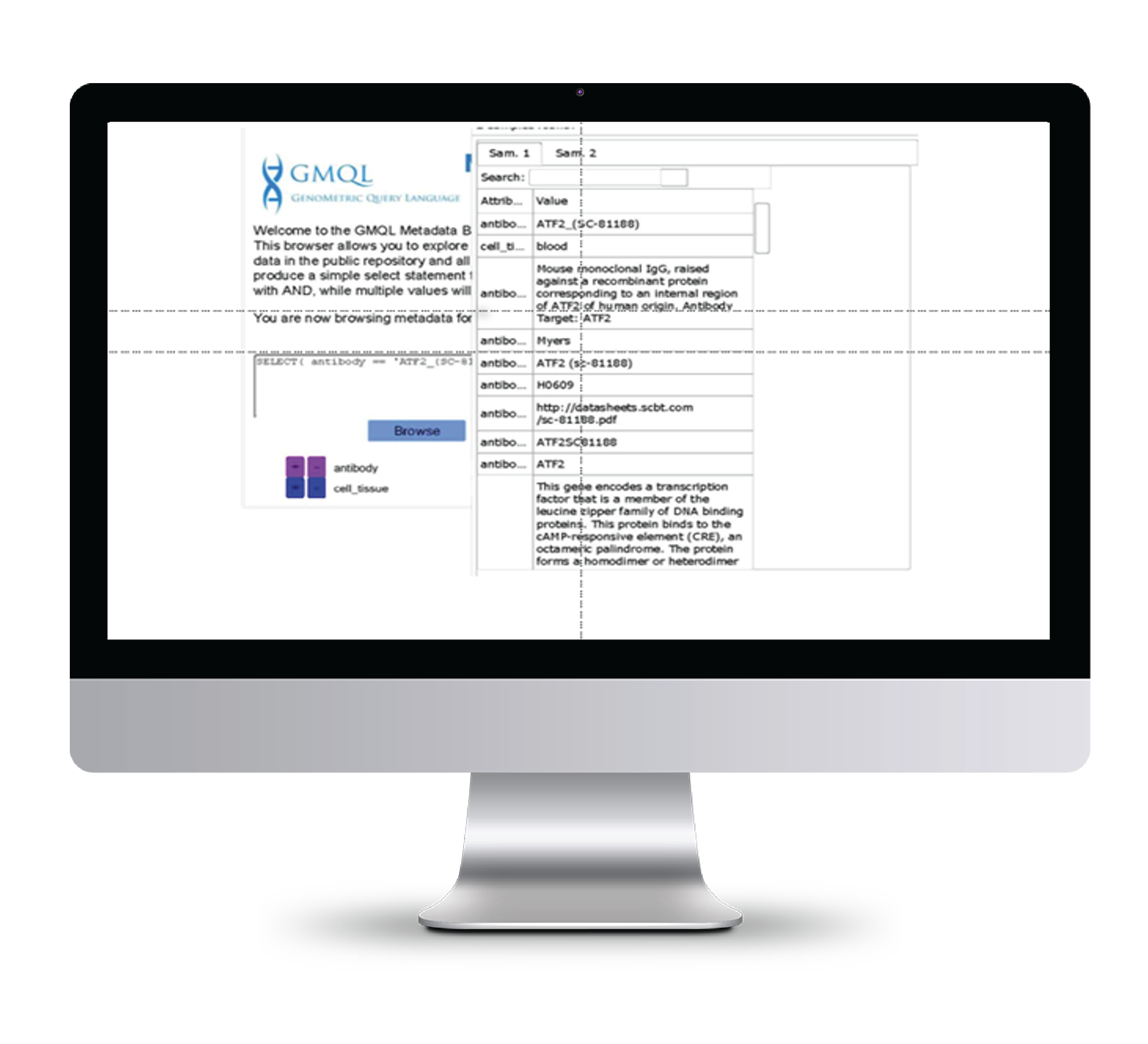
Genome space analysis
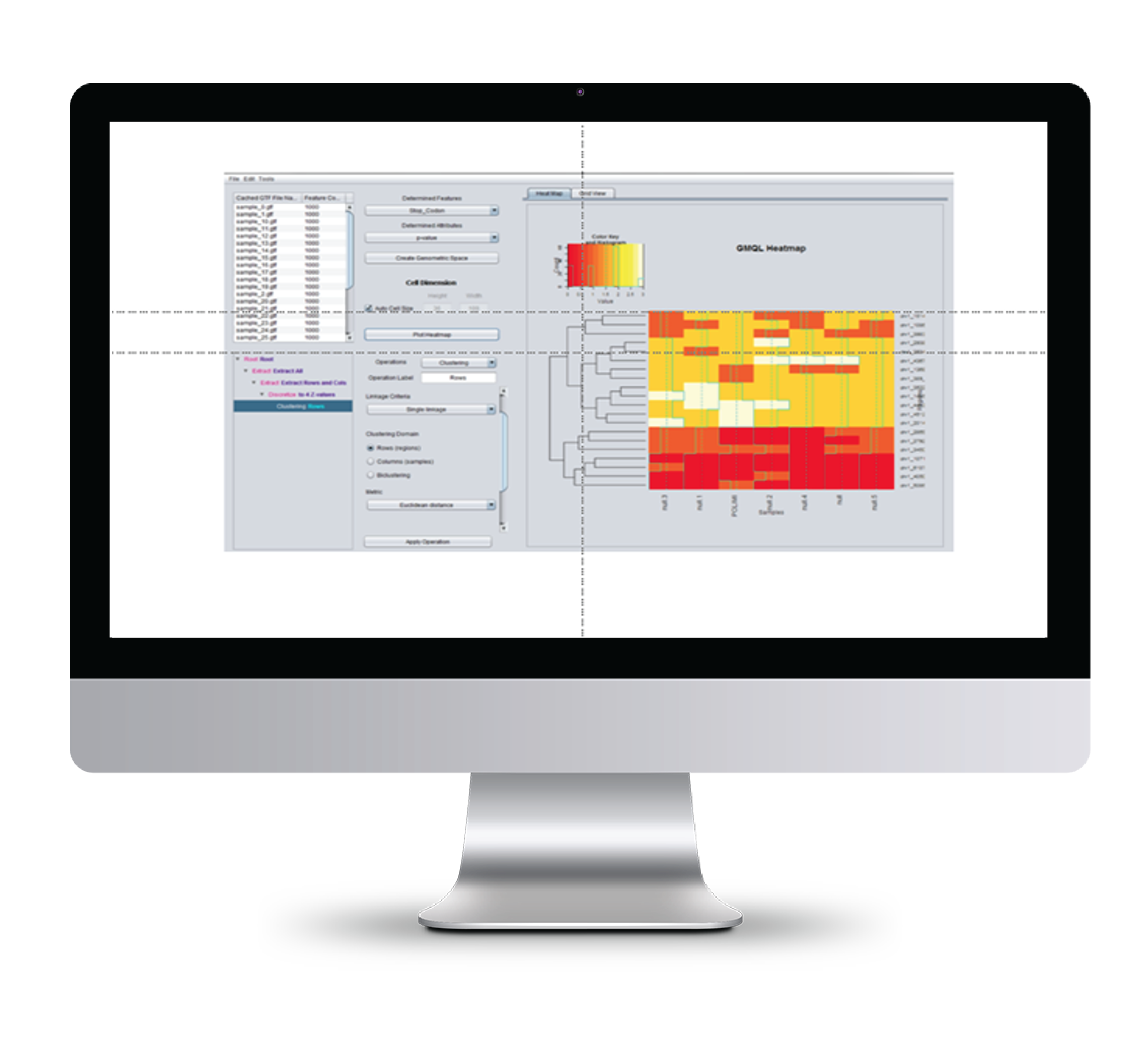
We also developed a output client-based tool for analyzing genome spaces, i.e. tables of region and sample data generated through GMQL. The tool supports heat map visualization, ordering, and clustering or bi-clustering.
Data-Driven Genomic Computing
GenData 2020 was a PRIN Project financed by MIUR (March 2013 - February 2016), coordinated by Politecnico di Milano, and involving 9 research centers in Italian universities (Politecnico di Milano, Università di Bergamo, Università di Milano, Università di Torino, Università di Bologna, Università di Roma 1, Università di Roma 3, Università di Salerna, Università della Calabria) for exploring the research problems in data-centered genomic computing.

Stefano Ceri
Professor
DEIB, Politecnico di Milano

Marco Masseroli
Associate Professor
DEIB, Politecnico di Milano

Vahid Jalili
Completed PhD cum Laude in 2015
DEIB, Politecnico di Milano

Abdulrahman Kaitoua
PhD Student
DEIB, Politecnico di Milano

Fernando Paluzzi
PhD Student
will complete PhD at IEO

Pietro Pinoli
PhD Student
DEIB, Politecnico di Milano

Yuriy Vaskin
PhD Student
will complete PhD at Novosibirsk State University

Francesco Venco
Completed PhD cum Laude in 2015
DEIB, Politecnico di Milano
Collaborators

Gianpaolo Cugola
Professor
DEIB, Politecnico di Milano

Matteo Matteucci
Associate Professor
DEIB, Politecnico di Milano

Heiko Muller
Senior Researcher
IIT@SEMM

Chiara Leonardi
Research Fellow
DEIB, Politecnico di Milano





Stefano Ceri: ceri AT elet DOT polimi DOT it
Marco Masseroli: masseroli AT elet DOT polimi DOT it
via Ponzio 34/5, Milan
+39 02 23993400