MyWEST configuration
Many parameters are available in MyWEST to optimize the mining algorithm performance for the specific extraction, change the database where storing the extracted data, and define new web databanks from which retrieving annotations. Values for all these parameters can be configured by editing the two text files:
This text file contains the default values for the extraction parameters. When a new template is created, also a file called "TemplateName_Config.txt" is created in the MyWEST/data/ directory. This new file, a copy of the default Config.txt file, is the default extraction configuration file for the created template and can be modified through the MyWEST graphical interface by clicking the Change button in the template building interface of MyWEST Template configuration panel (Figure 1).
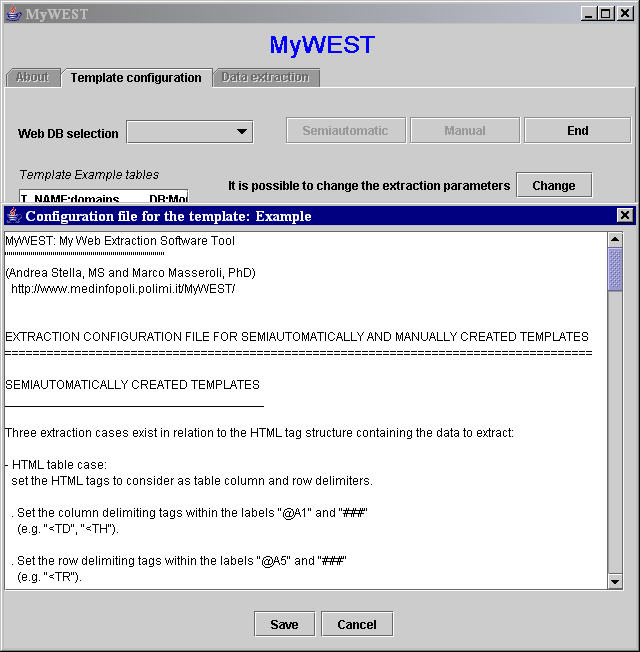 |
| Figure 1. Graphical interface for modifying the template extraction configuration file. |
In the Config.txt and TemplateName_Config.txt files, extraction parameters are grouped according to the function type they refer to. Each parameter is identified by a specific tag, and the list of set values of each parameter is delimited at the beginning by the parameter tag and at the end by the "###" tag. Parameter tags have the form "@Xy" (e.g. @A3), where X is the function type to which the parameter refers (i.e. A for functions used with semiAutomatically created templates, M for functions used with Manually created templates, and C for functions used for extracted data Cleaning), and y is the incremental parameter ID in each group.
Parameters @A1 - @A5 and @M1 - @M2 enable to optimize extraction performance of semiautomatically and manually created templates, respectively.
Parameters @C1 - @C2 allow cleaning extracted data by removing from the beginning and the end of each extracted information the list of characters specified as values of the @C1 and @C2 parameters, respectively.
- Example 1
- In order to extract the H.sapiens protein similarity information from the UniGene database HTML page in Figure 2, as explained in the Basic User Guide (1) section, it can be used a template semiautomatically created providing the sequence of characters SIMILARITIES as anchor, and the two sequences of characters H.sapiens and asparagine to identify the HTML structure containing the data to extract from the page.
Because in this case the data to extract are contained in a HTML table structure, during extractions the @A1 and @A5 parameters are considered and extraction performance depends on the @A1 and @A5 parameter values set in the TemplateName_Config.txt file. In fact, parameters @A1 and @A5 indicate the HTML tags to consider as HTML table column and table row delimiters, respectively.
In the UniGene database HTML page in Figure 2, the HTML table structure containing the data to extract has columns and rows delimited by the <TD> and <TR> tags, respectively. Thus, for extracting the data structured as they appear formatted in the HTML page, set the @A1 and @A5 parameter values to <TD and <TR, respectively. Generally, leaving the @A1 and @A5 parameters set to their default values (i.e. <TD and <TH for @A1, and <TR for @A5) makes the application extracting the data of interest correctly.
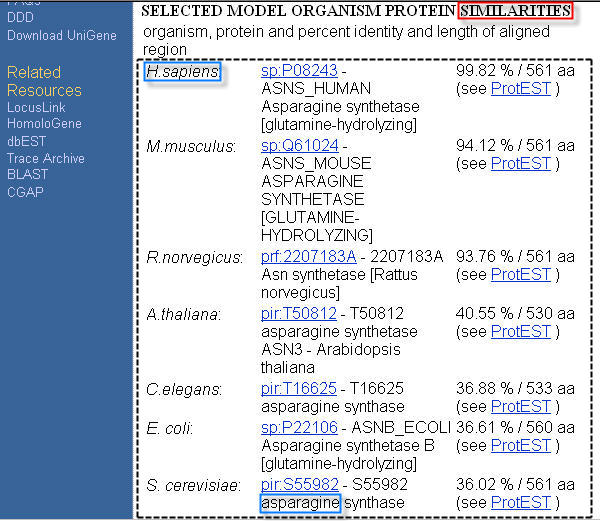 |
| Figure 2. Example of HTML page from the UniGene database. |
- Example 2
- To extract the Protein domains annotation from the Mouse Genome Informatics database HTML page in Figure 3, as explained in the Basic User Guide (1) section, it can be used a template semiautomatically created providing the sequence of characters domains as anchor, and the two sequences of characters Description and Neurotransmitter-gated ion-channel ligand binding domain to identify the HTML structure containing the data to extract from the page.
In this case, the data to extract are contained in a HTML table structure. Therefore, during extractions the @A1 and @A5 parameters are considered and extraction performance depends on the @A1 and @A5 parameter values set in the TemplateName_Config.txt file. In fact, parameters @A1 and @A5 indicate the HTML tags to consider as HTML table column and table row delimiters, respectively.
In the Mouse Genome Informatics database HTML page in Figure 3, the HTML table structure containing the data to extract has columns and rows delimited by the <TD> and <TR> tags, respectively. Thus, for extracting the data structured as they appear formatted in the HTML page, set the @A1 and @A5 parameter values to <TD and <TR, respectively. Thus, leaving the @A1 and @A5 parameters set to their default values (i.e. <TD and <TH for @A1, and <TR for @A5) makes the application extracting the data of interest correctly.
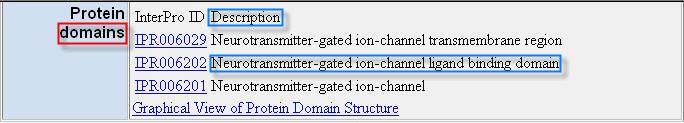 |
| Figure 3. “Protein domains” annotations for the MGI:87895 gene in the correspondent HTML page of the Mouse Genome Informatics (MGI) database. |
- Example 3
- In order to extract the M.musculus protein similarity information from the UniGene database HTML page in Figure 2, as explained in the Basic User Guide (1) section, it can be used a template manually created providing the sequence of characters M.musculus as anchor, and the sequence of characters SP:Q61751 to identify the HTML structure containing the data to extract from the page.
During extractions, the @M1 and @M2 parameters are considered and extraction performance depends on the @M1 and @M2 parameter values set in the TemplateName_Config.txt file. @M1 can assume value <REDUCED or <EXTENDED. <REDUCED indicates that only the characters delimited by the same HTML tag delimiting the second sequence of characters provided during the manual template creation (i.e. SP:Q61751) will be extracted. <EXTENDED indicates that also neighbor characters delimited by the closest of the HTML tags set as values of the @M2 parameter will be extracted.
Thus:- if @M1 is set to <REDUCED, only the “SP:Q61751” sequence of characters is extracted from the HTML page in Figure 2 because it is included in the same link HTML tag delimiting the sequence of characters selected, during the template creation, to automatically identify the HTML structure containing the data to extract. (In fact, in the HLML page in Figure 2 the “SP:Q61751” sequence of characters appears with the same style, different from its neighbor characters, of the sequence of characters selected during the template creation to automatically identify the HTML structure containing the data to extract);
- if @M1 is set to <EXTENDED:
- if <TD is among the values set for @M2, the sequence “SP:Q61751 - TC17 MOUSE TRANSCRIPTION FACTOR 17” is extracted because in the HTML page in Figure 2 this sequence of characters is delimited by the <TD tag;
- if <TR is among the values set for @M2 and <TD is not, the sequence “M.musculus: SP:Q61751 - TC17 MOUSE TRANSCRIPTION FACTOR 17 45 % 54 aa” is extracted because in the HTML page in Figure 2 this sequence of characters is delimited by the <TR tag.
- if <TD is among the values set for @M2, the sequence “SP:Q61751 - TC17 MOUSE TRANSCRIPTION FACTOR 17” is extracted because in the HTML page in Figure 2 this sequence of characters is delimited by the <TD tag;
- if @M1 is set to <REDUCED, only the “SP:Q61751” sequence of characters is extracted from the HTML page in Figure 2 because it is included in the same link HTML tag delimiting the sequence of characters selected, during the template creation, to automatically identify the HTML structure containing the data to extract. (In fact, in the HLML page in Figure 2 the “SP:Q61751” sequence of characters appears with the same style, different from its neighbor characters, of the sequence of characters selected during the template creation to automatically identify the HTML structure containing the data to extract);
In this file are stored the configuration parameters used to define either linking connections to the web databanks from which retrieving the HTML pages containing the data to extract, or the connection to the database where storing the extracted annotations. The DatabasesURL.txt file can be modified through the MyWEST graphical interface by clicking the Modify Web DBs button in the MyWEST Template configuration panel (Figure 4).
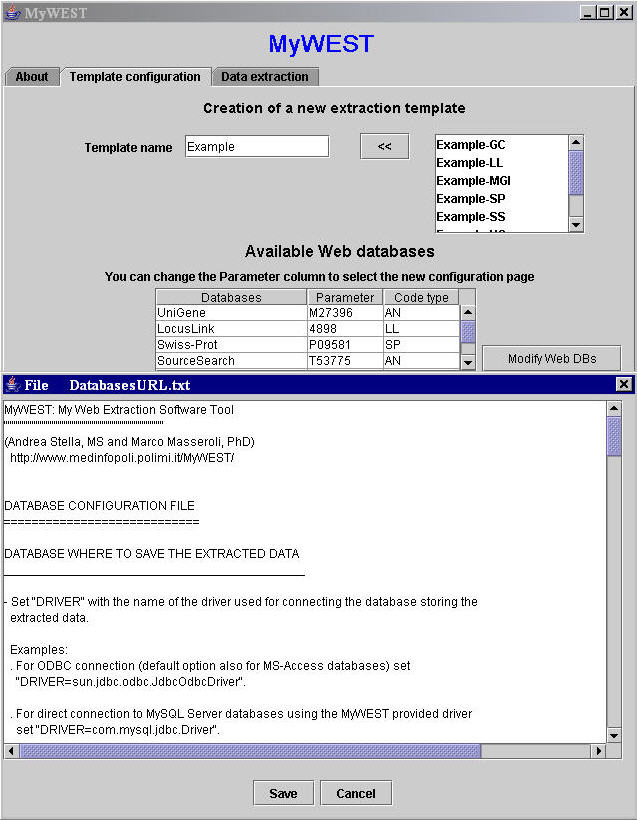 |
| Figure 4. Graphical interface for modifying database connection parameter values. |
The parameters used for identifying and accessing the database where storing the extracted data are:
- DRIVER, DBURL, USER, PASSWORD
Any kind of database with a suitable schema can be used. The database can reside locally in the used computer or remotely. Details on parameter value setting and on how using specific drivers for database connection, can be found here and in the DatabasesURL.txt file.
For accessing Web databases through Internet, it is possible to use a proxy server. In this case, the PROXYSERVER and PORT parameters must be set with the proxy IP number or name, and proxy port number, respectively. Otherwise, to not use a proxy server connection, set "PROXYSERVER=NOPROXY" and "PORT=".
The parameters used to connect to a web database and retrieve its HTML pages are:
- NAME, MODE, BASEURL, PARAMETER, URL1, LINK, CODE_TYPE
The URL1 and LINK parameters must be used when a web database server requested for a specific nucleotide or aminoacidic sequence code does not provide directly the HTML page containing the annotations of the specified sequence. Instead it provides an intermediate HTML page containing a link to the requested HTML page.
Refer to the DatabasesURL.txt file for details on possible parameter values and on how setting new parameter values for a new web database to extract the data from.
- Example
- Three examples of parameter settings configured to access and automatically retrieve HTML pages from the LocusLink, GeneCards and Mouse Genome Informatics web databanks, respectively, are following presented.
- NAME=LocusLink
BASEURL=http://www.ncbi.nlm.nih.gov/LocusLink/LocRpt.cgi?l=
PARAMETER=4898
CODE_TYPE=LL
END
- NAME=GeneCards
BASEURL=http://bioinfo.weizmann.ac.il/cards-bin/cardsearch.pl?search=
PARAMETER=AA576511
LINK=Display
URL1=http://bioinfo.weizmann.ac.il/cards-bin/
CODE_TYPE=AN
END
- NAME=Mouse Genome Informatics (MGI)
BASEURL=http://www.informatics.jax.org/searches/accession_report.cgi?id=
PARAMETER=MGI:87895
CODE_TYPE=MGI
END