Extraction from multiple HTML pages (here for a tutorial)
The MyWEST Data extraction panel (Figure 1) enables using the created templates to automatically extract annotations of many genes and/or proteins at a time in a batch mode from multiple HTML pages also of distinct web databanks.
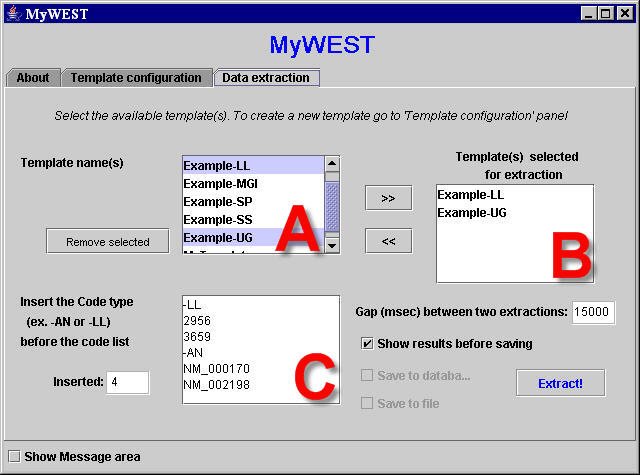 |
| Figure 1. "Data extraction" panel for the automatic batch extraction of manifold gene annotations from multiple HTML pages, also of distinct web biomolecular databanks, using the created templates. |
- The Data extraction panel allows defining which templates to use for the batch extraction of annotations of what genes/proteins. It also enables choosing whether visualize the extracted data or directly storing them and in which format.
- contains the list of created templates available to be used for defining which data to extract. The Remove selected button delete the selected template(s) for good!;
- contains the list of templates selected for the extraction. Templates can be added to this list by selecting them in A and clicking the “>>“ button; each template can be removed from the list by selecting it and clicking the “<<“ button;
- contains the list of nucleotide and/or aminoacidic sequence identification codes defining the HTML pages of a genomic databank that can contain the data to extract. In this text area, the identification codes must be inserted each in a distinct row and grouped in blocks according to their code type. Each block must be preceded by a row identifying the block code type and containing the same code type label associated in the DatabaseURL.txt file to the databank to extract the data from and for which the used template has been created (e.g. -AN for GenBank Accession Number, -LL for LocusLink ID, -SP for Swiss-Prot Accession Numbers).
The Gap between two extractions (msec) text field shows the gap in milliseconds (default 1000) between the data extractions of two subsequent sequence identification codes in the list in C. For long sequence identification code lists it is useful to set this value to 15000, at least, to avoid loading a databank web server with many concentrated requests. In many case this can lead to the permanent cutting of the connection to the web sever for the used computer IP.
When the Show results before saving checkbox is selected, after completing the extraction, result tables are shown for visual evaluation to the user who can decide if saving the mined data (Figure 2).
If the Show results before saving checkbox is not selected (extracted data are directly stored), the Save to database and Save to file checkboxes allow choosing in which format saving the extracted data (in a relational database and/or in text files).
The Extract! button makes the extraction beginning.
Actions performed by MyWEST during the extraction, together with messages reporting extraction successes and problems eventually occurred, are displayed in the Message area window. The same messages are stored in the extraction log file too.
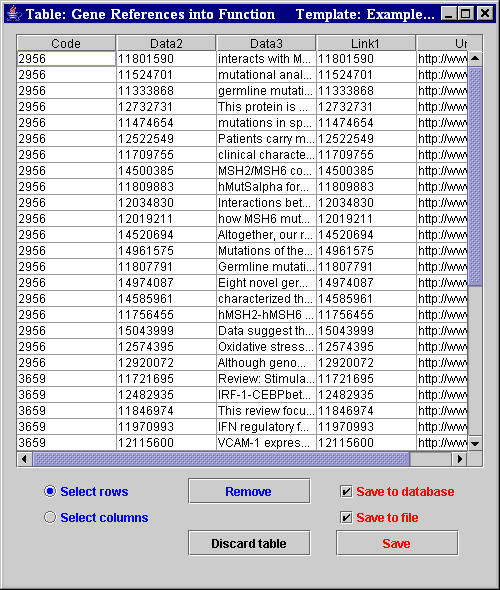 |
Figure 2. Result table after a batch Extraction. |
At the end of the extraction, if the checkbox Show results before
saving was set, an extraction result table (Figure 2) appears for each table defined in the used template. The user can visually evaluate the extracted annotations and choose whether discarding or saving the extracted data. It is also possible to manually filter the extraction results by selecting and removing columns and/or rows eventually containing useless data.
To rapidly evaluate extraction results, users can check the extraction log
files in the MyWEST/logs/ directory. These logs are Excel text files where each row contains information of a single performed data extraction, including extraction characteristics and short messages reporting extraction problems or successes (Figure 3).
Ordering the log file content by Message and Code, for each selected nucleotide and/or aminoacidic sequence, not extracted type of data and no extraction reason messages can be grouped, highlighting eventual extraction problems. This allows also to immediately evaluate completeness and goodness of a performed data extraction.
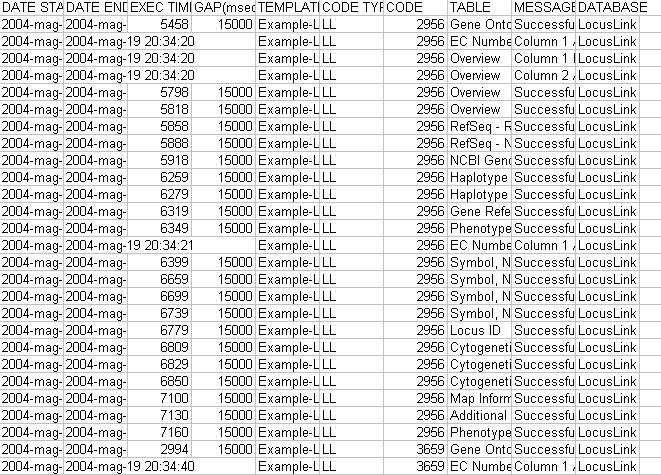 |
Figure 3. Example of an extraction log file. |
Annotations extracted from biomolecular databanks with high temporal variability can be automatically updated using MyWEST updating module after connection to a relational database has been correctly configured.
To this aim, run the file MyWEST-Up.bat in the MyWEST/updating directory after editing it to configure the command line:
javaw -jar MyWEST-Up.jar TEMPLATE_LIST CODE_LIST gap=GAP output=SAVING_TYPEby assigning a value to the TEMPLATE_LIST, CODE_LIST, GAP, and SAVING_TYPE parameters (e.g. javaw -jar MyWEST-Up.jar updating/Templates.txt datafiles/Codes.txt gap=15000 output=xls+db) according to the following:
- TEMPLATE_LIST: name, with extension and relative path from the main MyWEST directory, of the text file containing the list of templates to use for the extraction (e.g. updating/Templates.txt).
-
CODE_LIST: name, with extension and relative path from the main MyWEST directory, of the text file containing the list of nucleotide and/or aminoacidic sequence codes to extract annotation for (e.g. updating/Codes.txt).
- GAP: interval in milliseconds between the data extractions of two subsequent sequence identification codes in code the list. For long sequence identification code lists it is useful to set this value to 15000, at least, to avoid loading a databank web server with many concentrated requests. In many case this can lead to the permanent cutting of the connection to the web sever for the used computer IP.
- SAVING_TYPE: saving format of the extracted data.
- xls: saving in excel text files;
- db: saving in a relational database;
- xls+db: saving both in excel text files and in a relational database.
- db: saving in a relational database;